The NFDI4Ing Base Service Research Software Development (S-2) provides infrastructure, best practices and templates to make research software and its development more replicable and reproducible while improving the quality of the written code. Currently, our main projects are a JupyterHub server for the NFDI4Ing-community, a knowledge base with best practices and examples for sustainable code development, and training courses for, e.g., GitLab.
Vision
Research software plays a fundamental role in the advancement of science. Not only is every simulation based on code and the assumptions in the models therein, but software also plays a key role in processing and evaluating almost every measurement. Therefore, the importance of functioning, quality-assured code and the effectiveness and effectivity of its development can hardly be over-emphasized. Our vision is a world in which every piece of code is well tested, quality-assured and easily reusable.
Reusable Workflows and Best practices
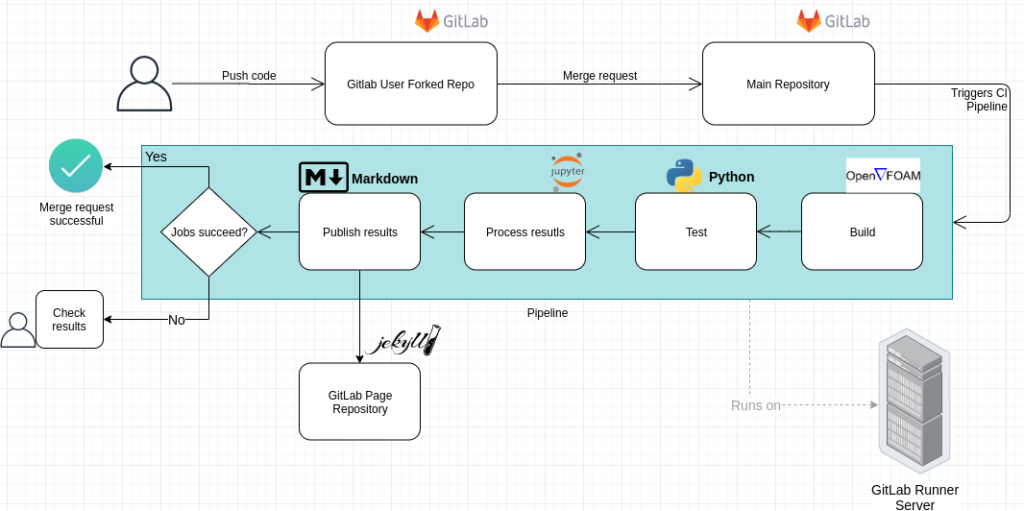
To help bring this vision to life, we provide and actively work on a knowledge base for software development (Link) as one of our services. Here we provide various guides and “How-to”-articles on different topics of research software development. The workflow proposed and the articles provided were developed and proven useful in the CRC 1194 at TU Darmstadt (Link), effectively combining version control, continuous integration, automatic testing and evaluation, including the particular requierments of HPC-systems. The Knowledge Base is an ongoing project, in which we are working together with participants in- and outside of NFDI4Ing. In 2022, we presented the Knowledge Base at the FOSDEM’22, a developers conference with thousands of participants. The presentation is accessible here (Link). For further information or if you want to contribute, please contact us (Link)! We always welcome new contributors!
Jupyter at your workstation
To support parallel code development and documentation, but also to make reproducing analyses easier, we have set up a JupyterHub that allows scientists to develop or reuse Jupyter notebooks. In these, code can be executed interactively and enriched with text, figures and more (i.e. literate programming). At the moment, the JupyterHub (Link) is still in the testing phase and not yet available to the public, but it will go live within this year.
One particular use case we are currently working on is integrating MATLAB® for academic usage. It is possible to work within the known MATLAB® IDE, create and run MATLAB® live
scripts, or to use a MATLAB® kernel within the classical Jupyterlab environment. Furthermore, polyglot notebooks, i.e. notebooks with different languages in each code cell, will be possible.
If these use cases or the JupyterHub in general sounds interesting to you, please contact us (Link) and become a test user!
Christian Bischof, Moritz Schwarzmeier, Anett Seeland